
GEORG URTEL in Bali
Young and inspired postdocs in science have a way of moving around pretty fast. It was several weeks before I heard from Georg Urtel in response to my request for an interview about a recent experiment at Ludwig Maximilians University, Munich on reversible switching of cooperating DNA hairpin replicators and a paper that he was “first author” of.
Urtel earlier this year left Dieter Braun’s lab at LMU — where his experiment was part of his PhD — for postdoctoral research at Laboratoire Jean Perrin, Pierre-and-Marie-Curie University, Paris. Urtel’s LMU collaborators on the experiment, Dieter Braun and Thomas Rind, were not available for comment. Rind following his time in Braun’s lab was recruited into industry.
Braun, in addition to the hats he wears as director of his systems biophysics lab at LMU and of his own origins of life group, serves as one of the investigators in the Simons Collaboration on Origins of Life. As a postdoc, he too did some fancy work in the lab of Rockefeller University physicist Albert Libchaber a dozen or so years ago, which Libchaber once described to me in a chat about origin of life:
“The little volcanoes that make thermal vents are full of porous material, and those pores hold all the temperature gradients you need. What we showed is that with this temperature gradient many nonequilibrium processes exist.
For example, we discovered that polymerase chain reaction (PCR) is possible under thermal conditions. There’s nothing but thermal convection. In the center region of a convective cell double-stranded (DS) DNA melts and in the side single-stranded (SS) DNA is copied. We thus made the smallest PCR machine, centimeter in size.”
PCR plays a central role in the recent Urtel et al. hairpins experiment at LMU.
Georg Urtel is a native of Austria (citizen of Austria and Germany). His BS in particle physics, MS in biophysics, and PhD in systems biophysics – magna cum laude -- are all from Ludwig Maximilians University. He’s been a guest scientist in Paris at CNRS’s Laboratoire de Photonique et de Nanostructures and at UPMC. He is co-author of “Synthesis and materialization of a reaction-diffusion French flag pattern” (Nature Chemistry, in press).
I spoke by phone with Georg Urtel at his home in Paris.
Suzan Mazur: In your recent nucleic acid sequences as replicators experiment, DNA hairpins were able to return to their original hairpin structures after creating a new type of replicator. I’m aware of the catalytic quality of stem-loop RNAs, but you worked with single stranded DNA. What gave the DNA its catalytic ability?
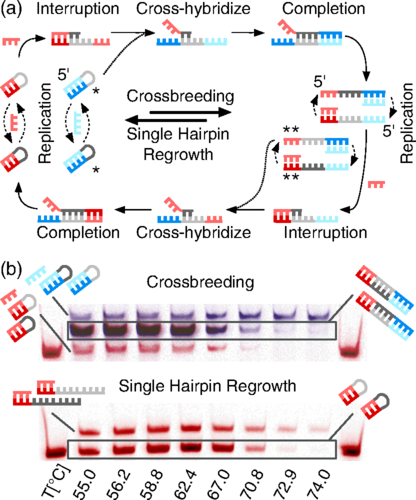
Georg Urtel: The DNA we used for the hairpins experiment did not have any catalytic activity at all. We used PCR, i.e., polymerase chain reaction as replication machinery. It’s kind of a highly evolved machinery, especially the one we used. It has proofreading function and so on.
But it’s not a lot of proxy, because we were more interested in how the secondary structure of the DNA has an effect on how it is replicated. It doesn’t really matter what replication machinery you use with this experiment. We also tried different polymerases. All showed the same result.
With RNA, this would just be a bit more tricky from the experimental side because you’d have to use RNA polymerases. Similar to the ones Gerald Joyce or Philipp Holliger is using. But with our experiment -- we made 60 rounds of replications and had secondary structures -- it probably wouldn’t have been feasible yet with RNA.
Suzan Mazur: So you’re saying the catalytic quality was solely due to the machinery, the PCR you were using.
Georg Urtel: Yes. Exactly. We looked at the secondary structures of the DNA strands and how they affect replication. But the replication was entirely done by the polymerase enzyme.
Suzan Mazur: Would you briefly describe the experiment? You added primers and buffer to the DNA polymerase and then you put the samples through temperature cycles.
George Urtel: We started with DNA, with hairpin structures. The hairpins are templates for replication. In order to be recognized by the DNA polymerase, the DNA templates need those primers. The primers are then elongated according to the template. But you don’t end up with the same strand of DNA, you end up with a complementary strand.
In a classical experiment, you’d need a second primer for this complementary strand, to form the exact copy of the initial strand.
If you think of a primordial soup or mixture, perhaps the sequence space was not very big and there was not a huge variety of sequences. Maybe only a few types of primers existed. That’s why the secondary structure of DNA is important to consider. When copying the hairpins, the complementary strand will have the same binding site for the primer. Thus, only one primer is needed for exponential replication.
Suzan Mazur: Aside from serving as templates, what else do the hairpins do in the experiment?
Georg Urtel: The hairpins are just being replicated. They are being replicated very inefficiently. Again, this has to do with the nature of the secondary structures. The hairpin structure is inhibiting because what the hairpin structure does is it forms a double helix with itself. When you have such a structure, the primer can’t bind.
Suzan Mazur: What would you say is the significance of this experiment? Hasn’t it been known for many years that hairpins have a role in replication and recombination?
Georg Urtel: Yes. The hairpin structure you find in all kinds of RNA enzymes, but the point here is that we started with a simple replicator, the hairpin, that only uses one primer. And although this results in very inefficient replication -- we’ve shown that two hairpins with some similarities in the loop structure can form a crossbreed that doesn’t have the secondary structure anymore. The crossbreed uses both primers and is replicating extremely fast.
Suzan Mazur: And no RNA is evident, only DNA.
Georg Urtel: Exactly. We work with DNA for various reasons. It’s easier to do an experiment with DNA. That’s also why we use the polymerase. What we were looking at in our experiment was the secondary structure.
In this regard RNA and DNA are very similar. Both are polymers with four different bases. In the case of DNA, they are often just called A, C, G and T. They form two complementary pairs: A-T and C-G. That means, if you write down a sequence, you can easily determine the complementary sequence required to bind to the sequence and form a double helix.
Suzan Mazur: Have other scientists done experiments along the lines of yours following your experiment?
Georg Urtel: I think there may have been experiments done, for example, in our group where PCRs (polymerase chain reactions) were used just as a proxy for any replication mechanism. However, the key mechanism is new to our reversible switching of cooperating replicators experiment.
The key mechanism is that you have an incomplete replication -- which is something that can always happen -- but in our particular experiment there is a benefit from the incomplete replication.
Suzan Mazur: You’re referring to regrowth of the original hairpin?
Georg Urtel: The incomplete replication is important in two ways. It helps make the transition from hairpin replicators to the fast crossbreeds, and when conditions are changed and only one primer is present, the same mechanism can be used to regrow the original hairpins. The crossbreed goes extinct under such conditions.
Suzan Mazur: So in the scheme of things, the origin of life picture – where does your experiment fit in?
Georg Urtel: The origin of life, of course, covers a very broad span. We are at a later stage. We did not combine chemicals to get amino acids and nucleic acids.
Suzan Mazur: When you refer to “auto- and cross-catalytic processes” in your paper, you’re not talking about autocatalytic sets.
Georg Urtel: We are not talking about autocatalytic sets. Autocatalysis for us is from the viewpoint of the primer. When the hairpin is copied, you always create something that is copied with the same primer. And when you have this cross-catalysis – the type of replication of the crossbreed -- then you have a species that uses two different primers. It’s making cross-catalysis in a way in which you need primer 1 to copy template 1, which will give you the complement of template 1. You need primer 2 to finish the job to go back to the initial template.
What I think is also interesting regarding our experiment is that the benefit is a very peculiar one because we have an information transfer happening from the hairpins to the crossbreed, but the actual hairpins in the experiment can completely vanish, go extinct. After the hairpins give their information to the crossbreed and vanish, we can then bring them back from the dead.
Suzan Mazur: Thank you. You used some language in your prebiotic evolution paper that reflects Darwinian science, terms like selection and fitness, even though you’re not actually doing Darwinian science. Many scientists now say the language of evolutionary science needs an overhaul. That the scientific language of the 19th century is not particularly accurate or productive anymore. I’m referring to Herbert Spencer’s famous reference to survival of the fittest: ”Nature, red in tooth and claw.” What are your thoughts?
Georg Urtel: I think it’s a difficult question. You want to be accurate, yes. But you also want to reach people. So sometimes a word may be used in a paper so people can more easily recognize a concept, although it may be more precise to use five words to describe the concept. I think the terms you cite in our paper are not outdated in that sense.
Suzan Mazur: Where do you think nucleic acid was first produced? What is its origin?
Georg Urtel: Actually I don’t care too much because what’s important is that we come up with approaches to the synthesis of life. It’s unlikely we will find THE route from chemicals to the first cell, as it originally happened.
The emergence of life from an RNA World is, of course, a popular hypothesis. But we just don’t know. Maybe there were several RNA Worlds, maybe there were several DNA Worlds that went extinct and maybe nucleic acids had different origins in these worlds. The situation on other planets is also an unfolding narrative. Exploring the possibilities is what’s important.
Suzan Mazur: Are you still working along these lines now in Paris?
Georg Urtel: I’m doing postdoctoral research at Pierre-and-Marie-Curie University here in Paris on biochemical reaction networks. I’m not currently working on origin of life.
Suzan Mazur: Is your hairpin structures as replicators experiment worth a followup or do you think you’ve made your point?
Georg Urtel: It would be good to do followups. The key mechanism is so interesting. It works with incomplete replication. It’s something you actually get for free. All you need is a process (like an increase in temperature) to interrupt the replication.
