I recently had a three-way phone conversation with Swedish deep evolution investigators Charles Kurland and Ajith Harish about their phylogenomic Tree of Life (ToL) based on protein structure, which shows that we are descended from a “complex” ancestor — MRUCA (most recent universal common ancestor) -- not a simple bacteria. Kurland and Harish think a ToL paradigm shift may be in order. What’s more, Kurland and Harish figure that MRUCA was not the first ancestor, but represents complex survivors of a now-extinct biosphere.
The findings of Kurland and Harish challenge not only mainstream ToL perspectives, but also those of endosymbiosis hypothesis fans, as well as the “HGT industry” — as Kurland describes the inflated role he sees assigned to lateral gene transfer in evolution.
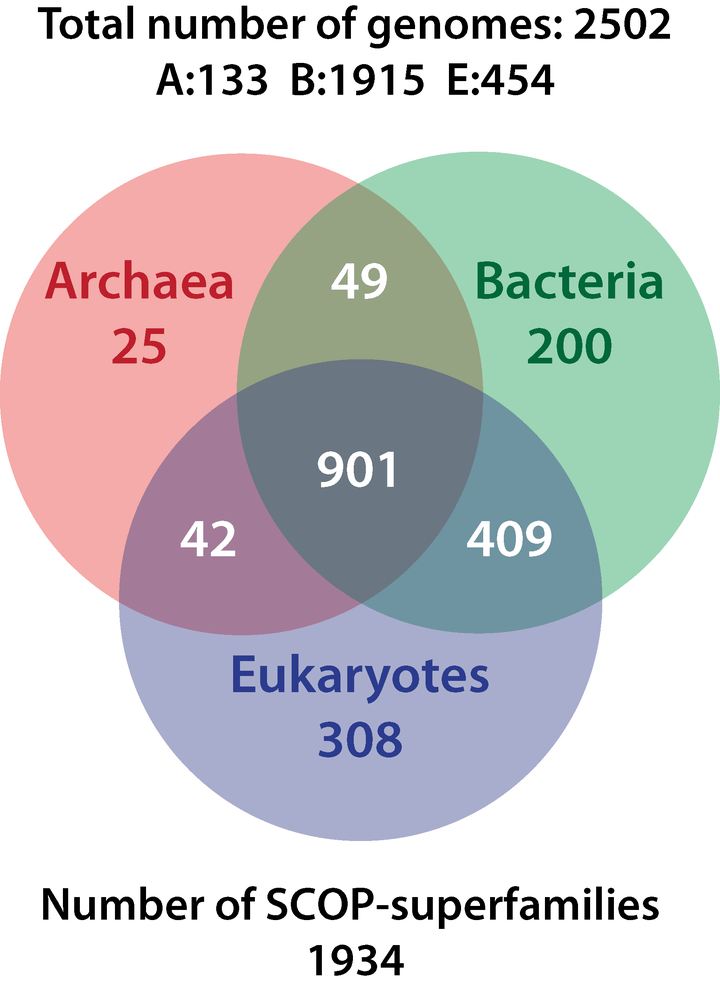
Kurland and Harish have published several scientific papers showing MRUCA shares three-quarters of its proteome with eukaryotes, bacteria and archaea (see Venn diagram above). They have another paper forthcoming, which they say reconfirms their results. Part of the investigation in recent years has been supported by the Nobel Committee for Chemistry of the Royal Swedish Academy of Sciences.
The Kurland-Harish research is computational but starts with experiments, i.e., x-ray crystallography of proteins, which are then translated to statistical models used to assign genome sequences to the experimentally determined structures.
Says Harish: “Our data contradicts the mainstream scenario(s) for origins of life as we know it, based on gene trees. Such hypotheses are based on analyses that are about two decades old and were artifact prone.”
Charles Kurland and Ajith Harish first developed a professional rapport at the University of Illinois, Urbana-Champaign more than a dozen years ago and were initially both very influenced by the work there of Carl Woese, the discoverer of archaea. Harish says he took a class at UIUC taught by Woese: “which got me into evolution.” But neither of them agrees now with the Woese Tree.
Kurland, a native New Yorker, loves Scandinavia and settled in Sweden decades ago, where he is now Professor Emeritus of Molecular Biology at both Lund University and Uppsala University and a member of the Royal Swedish Academy of Sciences, Stockholm. He is also a member of the Royal Science Society, Uppsala; the Royal Academy of Sciences, Copenhagen; the Estonian Academy of Science, Tallin; the Royal Physiographical Society, Lund; and the European Molecular Biology Organization.
Kurland’s PhD is in biochemistry from Harvard, where he was part of James Watson’s team in the early 1960s. He is known for his study of the E. coli ribosome. But also for his talent as a lively and highly effective communicator of science.
Ajith Harish, is a native of Bangalore, India, where he received his BS (St. Joseph’s College -- microbiology, chemistry and botany) and MS (Bangalore University -- biotechnology). Harish says his roots are in experimental science, studying plant molecular biology and physiology, “in vitro” and “in vivo,” but that his analyses are now “in silico,” computational.
After receiving his PhD (2010) from UIUC in biology (bioinformatics and molecular evolution), Harish headed north, to Sweden, for postdoc research at Lund University and to continue his collaboration with Charles Kurland. Harish has also been a researcher at Uppsala University in recent years, working in the lab of molecular biologist Mans Ehrenberg. He is currently a Guest Researcher at Uppsala.
Part 1 of my interview with Charles Kurland and Ajith Harish follows. Part 2 of our discussion (coming up) looks at how their ToL analysis further challenges today’s science.
Suzan Mazur: Charles, are you a native New Yorker? I detect some accent.
Charles Kurland: Yes, I was born in the Bronx.
Suzan Mazur: Are you a graduate of Bronx High School of Science?
Charles Kurland: No, I went to Great Neck High School because by that time my parents had moved to Long Island.
Suzan Mazur: You’re a member of the Royal Swedish Academy of Sciences now, no longer a Foreign Member -- does that mean you are a citizen of Sweden?
Charles Kurland: I’m a dual citizen of Sweden and the United States.
Suzan Mazur: Ajith, I know you are a native of Bangalore and that you received your BS and MS from schools in Bangalore, but for many years you’ve been working in Sweden, sometimes in Mans Ehrenberg’s lab at Uppsala. Do you also have dual citizenship?

Ajith Harish: I’m a citizen of India and a permanent resident of Sweden with something like a US green card.
Suzan Mazur: Charles, would you establish yours and Ajith’s central position regarding the Tree of Life?
Charles Kurland: The basic idea is -- and I think Ajith and I are rather convinced of this now – something like the Tree of Life that Ernst Mayr was talking about in his argument with Carl Woese. In 1998, there were two perspective papers published in PNAS. One was an attack by Mayr on what he considered a very “unbiological” Tree of Life, which was the Woese Tree of Life, and Woese’s counterattack.
As it turns out, all this was for technical reasons that were wrong. That was very well established by French investigators like Forterre and Philippe, who did a great Yeoman’s service in cleaning up the effects of biased mutation rates on gene trees.
Both Mayr and Woese were kind of talking from different points of view. Carl was defending – and he always was a champion – genotypic views of evolution, sequence-based. Sequence for him was the whole business.
Mayr all he wanted, being really a biologist – you can say more a naturalist than an evolutionary biologist. Maybe that’s unfair, I don’t know. But Mayr was very much a biologist and very much into using phenotypes of organisms for his understanding of phylogeny.
Those, unfortunately, were two very opposed points of view.
Suzan Mazur: When you say the French work on TOL, are you referring to early or more recent?
Charles Kurland: The French started on this in the 1980s. Carl just ignored them. Everybody just ignored them.
There is a tendency — and this is nothing special to the United States – but the scientific community in the United States is a community and it tends to insulate itself from other scientific communities.
Woese did tremendous work establishing the genotypic or sequence-based way of doing phylogeny. Tremendously important. He was certainly a major pioneer in this. The problem was that he did not himself realize that he was skating on thin ice because there was an artifact (a bias) buried in his work that completely ruined his phylogenies.
That artifact was the one that was very well worked out by Forterre and his people and by Philippe in the 1980s and 1990s and then into the first few years of this century. They and their collaborators – names like Delsuc and Brinkmann and so on, really did a bang-up job showing that gene trees are very susceptible to biased mutation rates.
In a gene tree you’re counting mutants and then you’re trying to plot some kind of rate of mutant accumulation as a rate of evolution. But if that rate is uneven, then you’re in lots of trouble. And, unfortunately, with the genes used in the Woese model it was vastly uneven.
The reason why people didn’t appreciate it early in the 70s or 80s was that everybody was very taken up by the view of neutrality, especially in the United States, where Jukes and King and certainly Kimura were the leading figures and had an enormous influence on population genetics.
Suzan Mazur: Neutrality is something Eugene Koonin has been talking about recently.
Charles Kurland: Unfortunately, a lot of people waited too long to look at it.
We are looking at phylogeny through the eyes of protein-domains, superfamilies, which have very well conserved structures that are selected structures. Therefore, the sequences that encode them are selected. This has been illustrated by studies of coding sequence variation by Dan Hartl’s group, they’ve illustrated that proteins that are under selection for structure and functions can change their sequences in a process they call “sequence meandering.” This is sequence drift, but to avoid confusion with neutral drift it is called meandering. In nature the superfamilies, which have a fixed structure, are represented by multiple coding sequences in the genome, and these sequences specify the same structures. They are intensely selected structures.
Suzan Mazur: Ajith, do you know if the University of Illinois, Urbana-Champaign – where you received your PhD in Molecular Evolution and Bioinformatics – and took a class with Carl Woese – is UIUC adhering to the Woese model?
Ajith Harish: I don’t know if anybody at UIUC has specifically moved from the Woese model. Probably not, but I’m not in touch with people there.
Suzan Mazur: Charles, can we go back to discussing the essence of yours and Ajith’s idea about the Tree of Life?
Charles Kurland: What Ajith and I believe, and I can assure you that neither one of us started out with this perspective, it’s something we learned from the data, which I’m now very proud of. That is because both of us had to change our ideas about things. I was friendly off and on with Carl Woese. He was a difficult fellow, but there were times that we were very close. And I was very much under his influence. And Ajith was very much under his influence by being a graduate student at UIUC.
Suzan Mazur: I did a feature interview with Carl Woese shortly before he died.
Charles Kurland: He was tough, but he could be a lot of fun. There were a lot of very good people at UIUC. Both Ajith and I were both very much under the influence of Carl. Our view emerged from the data after a lot of very painful experiments trying to make a sensible phylogeny. We found that there were three superkingdoms, but they were not arranged as Carl had said. They were arranged the way Ernst Mayr said they were arranged.
Ernst Mayr was very impressed with the cellular phenotype of eukaryotes as opposed to the cellular phenotypes of archaea and bacteria, which for him were just dots, very undeveloped images. The nucleus, the endoplasmic reticulum, the membranous systems – all of that for Ernst Mayr was a coherent phenotype for eukaryotes. It was clearly different from archaea and bacteria. And what struck Mayr was that archaea and bacteria are so similar to each other and different from eukaryotes. So Woese’s phylogeny with archaea and eukaryotes as sister clades made no sense to Mayr. That was the basis of his phenotypic argument. Of course, he made it in a much more sophisticated way.
What Ajith and I discovered was that yes, indeed, the phylogeny that we can see from a very, very thorough examination of genomic data, rather than a gene tree, is a common ancestor out of which two empires grow. One of them contains four or six major eukaryotic groups and the other one contains two major -- what they used to call “prokaryotes.” We prefer to call them “akaryotes” following Forterre because this notion of “pro” – that they came first – is definitely contradicted by the phylogeny we see. They seem to be “parallel” rather than “pro.” So we call them akaryotes, non-nuclear as opposed to the nucleated eukaryotes.
We’ve gone through test after test and this phylogeny holds.
Suzan Mazur: You discovered that of these three groups, there’s a commonality.
Charles Kurland: There’s a commonality between archaea, bacteria and eukaryotes. The universal common ancestor is the root of eukaryote and akaryote lineages and it contains more than a thousand superfamilies!
The other really remarkable thing, the thing that really stunned us is that there were people before us who had seen this also and at the time were totally ignored. The people I have in mind are Christos Ouzounis and his collaborators from Cambridge. They recognized that the universal common ancestor was a very complex ancestor. It is apparently a very big genome. But, now I have to qualify that. We cannot say more than that the universal common ancestor is a population. We don’t know how it is structured. We have no idea how many species/populations are in that ancestor.
Suzan Mazur: Ajith, can you explain a bit about your methods at arriving at the results and tell me how much commonality you found to the three groups?
Ajith Harish: We were looking at genomes and also we were looking at the Superfamilies, which are based on the three-dimensional structure. We have four nucleotides, 20 amino acids, 2,000 Superfamilies. These are the building blocks of both genes and genomes.
Suzan Mazur: What I’ve read in your papers is this. You’re saying eukaryotes and bacteria and archaea all descended from a most recent universal common ancestor that shared three-quarters of the Superfamilies of all three of these domains. That the Superfamilies are “homologous to those of modern mitochondria, chloroplasts, nuclei, spliceosomes, cytoskeletons and the like” and that this was “evident in the proteome of the most recent universal common ancestor as well as thinly dispersed among the eukaryotes.” That this ancestor represents complex survivors from a now extinct biosphere.
Ajith Harish: Correct. This is from phylogenetic reconstruction. When we do a phylogenetic reconstruction, we can trace back three quarters to a common ancestor. This is basically the genetic toolkit of that ancestral population.
Suzan Mazur: But how did you do this, what were your materials and methods? You say you used the Superfamily database. What is that?
Ajith Harish: There is something called the SCOP database, which is a structural classification of proteins. That’s where things start. In that hierarchy we have what are called Superfamilies, members of which can be inferred to be homologous. If you have many sequences in that Superfamily, all of them have a common ancestor.
Suzan Mazur: Where do you get the database?
Ajith Harish: SUPERFAMILY database takes these structural quantifications and annotates complete genomes.
Suzan Mazur: Who put together the database?
Ajith Harish: Julian Gough [British bioinformaticist].
Suzan Mazur: This is something that many researchers use?
Ajith Harish: Yes, this is publicly available.
Suzan Mazur: You also identified recurrent protein domains based on the Hidden Markov Models.
Ajith Harish: We have the Protein Data Bank (PDB), which has all the three-dimensional crystal structures. Starting from there you can assemble a sequence profile. You can use that sequence profile to identify all the homologs. This is done by the SUPERFAMILY database.
Suzan Mazur: This is based on x-ray crystallography.
Ajith Harish: You start with protein sequences for which the structure is known and then you use that information to build. You can find related sequences and build a probabilistic model. That’s what the Hidden Markov Model is. It’s a probabilistic model that can identify the protein structure in any given sequence that is related.
Suzan Mazur: What you’re doing, isn’t everyone doing that? What makes your approach unique?
Charles Kurland: No one is doing it.
Ajith Harish: Not many people use Superfamilies and genomes to build phylogenies. Most people use just gene sequences. Mainstream papers are based on 30 or 40 genes that we call the genealogy-defining core of genes. A majority of people construct gene trees based on gene sequences and what we are doing is comparing genomes. We are presenting as much of the genome as possible using a statistical protein structure model – Hidden Markov Model.
Charles Kurland: This notion of using the Superfamilies as an operational phylogenetic character. It comes out of a fantastic paper that was published in 1992 by Cyrus Chothia. He’s a bioinformaticist who’s been working at the MRC (Medical Research Council) in Cambridge. What he did was recognize that these folds made up a kind of alphabet of all the proteins on the planet. And he realized that there weren’t so many of them: Maybe a thousand. It turns out there are between two and three thousand, by present estimates.
What was very special about what Ajith and I did was to take this up very seriously and say, yes, let’s use this alphabet to do our phylogeny instead of nucleotides or amino acids. And it worked very, very well. We got the kinks out of it.
Suzan Mazur: Key geologists I’ve interviewed – particularly, Roger Buick, Bob Hazen and Mark McMenamin recognize the Snowball Earth scenario, i.e., global ice age, that is central to your thinking on deep evolution. There were several Snowball Earths, according to Bob Hazen. Some scientists think these global ice ages were precipitated by bacteria producing oxygen that destroyed the methane layer in the atmosphere. Buick’s research shows eukaryotes survived Snowball Earth, apparently. And McMenamin, who’s an expert on ediacara, also thinks eukaryotes survived Snowball Earth.
You’re saying something more. You’re saying eukaryotes and bacteria and archaea all descended from a common ancestor that shared three-quarters of the Superfamilies of all three of these domains. Charles, would you say more about the Snowball Earth scenario and the most recent universal common ancestor as you and Ajith see it?
Charles Kurland: It’s not simply Snowball Earth. It’s Snowball Earth that recovers in a volcanic period where temperatures go off-scale. It’s not clear what the crunch is, whether it’s the Snowball ice, per se, or what some people say is a Slushball. It’s not necessarily the low temperatures that are killing off the preexisting organisms. It can be the recovery. The volcanism can drive the temperatures up so high that much on this surface cannot survive.
The other thing is this. If you look at the fossil remains, at the micrographs that the paleontologists can produce -- it’s very interesting. They have by microscopic examination what they very tentatively identify as bacterial groups. But actually identifying bacteria in rocks as fossils is a tough game, and I think that none of those identifications are worth the paper they’re printed on.
Suzan Mazur: That’s fascinating.
Charles Kurland: All paleontologists have are some kind of little spherical, little microscopic something or others – the identity of which they don’t know. But they can find organic matter associated with these, so there is evidence of masses of very small, microscopic something or others and I wouldn’t call them bacteria. I wouldn’t call them archaea. We don’t know what they are.
We have to be very serious about this because a lot of early phylogenetic theory about archaea together with bacteria make eukaryotes, etc., is based on this language that presupposes we know what came first. And we don’t know what came first. All we know is that there are these little dots in micrographs.
The other thing that we know and this is really very interesting, particularly because of the results that Ajith and I got for the ancestor. There’s a history and it’s a very well documented history of big, round something or others, a thousand times larger than any bacteria. From the cellular point of view these are gigantic. And they have been seen as far back as 2.9 billion years ago, I read, perhaps 3.4 billion years ago.
Ajith Harish: 3.2 billion.
Charles Kurland: Okay, 3.2 billion years ago. And they’re called acritarchs. What they look like is something that we actually would identify with our big, fat common ancestor.
Suzan Mazur: How amazing.
Charles Kurland: Really intriguing. There’s not much science to go on it, it’s more aesthetic identification.
Suzan Mazur: Where have the acritarchs been found?
Charles Kurland: Everywhere. They were found in lots of different places and in lots of different ages and suddenly they disappear just before the Cambrian Explosion. And that’s when we expect the most recent universal common ancestor to take off -- in the Cambrian Explosion. Because we think that our phylogeny is essentially a description of the Cambrian Explosion.
Ajith Harish: Going back to Snowball Earth, the big issue is that we cannot really date the age of MRUCA (most recent universal common ancestor) because we don’t have any particular events to pinpoint it. Ours is a best case scenario. That is the limitation of all phylogenetic analysis. So we don’t really understand how other scientists can date the ancestor to 4 billion or 3 billion years.
