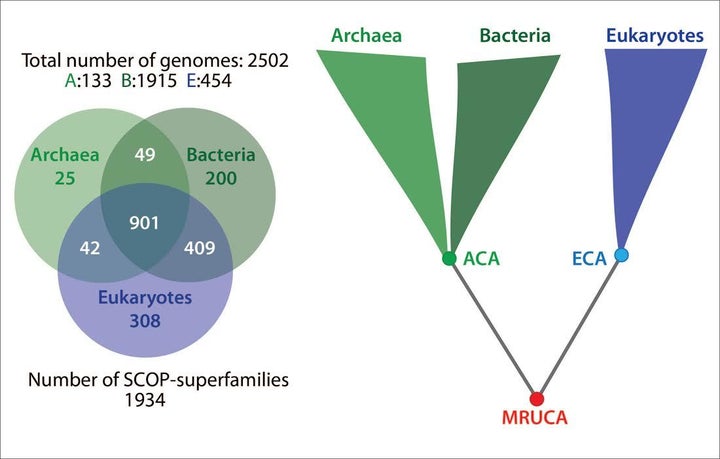
Part 2 of my conversation follows with Swedish deep evolution investigators Charles Kurland and Ajith Harish. We talk about how their Tree of Life — built with Superfamilies, genomes and three-dimensional crystal structures from the Protein Data Bank — challenges current thinking in evolutionary science (figure: (l) diagram showing Superfamily commonality, (r) Tree of Life).
See Part 1 of our interview, linked here, for biographical sketches of Charles Kurland and Ajith Harish and a discussion of their central position on deep evolution, which is that the most recent universal common ancestor (MRUCA) is complex not a simple bacteria and “is the root of eukaryote and akaryote lineages” containing “more than a thousand Superfamilies.” Kurland and Harish think MRUCA represents complex survivors from a now extinct biosphere.
Suzan Mazur: Your Tree of Life analysis challenges not only the work of Carl Woese and Lynn Margulis but John Maynard Smith, Ford Doolittle and Eugene Koonin, among others. You see endosymbiosis as wrong, and rampant horizontal gene transfer as a reflection of the “HGT Industry.” You think HGT was a negligible factor in evolution. Charles, would you address these two points, endosymbiosis and HGT?
Charles Kurland: Sure. They’re separate.
Suzan Mazur: I’d like to add one more point. I did an interview recently with Jeff Errington who’s investigating wall-less organisms called L-forms. In our conversation I referred to what you and Ajith are doing and your position on HGT and Jeff Errington said this:
“I think it’s likely HGT was fairly rampant in the primordial L-form-like cells. However, my feeling is that invention of the cell wall (perhaps separately in archaea and bacteria – which have completely distinct cell wall structure) could have brought an abrupt halt to the primordial HGT.”
Charles Kurland: Yes, absolutely. I agree with that.
Suzan Mazur: Errington said further:
“Once the wall was in place, new mechanisms of HGT may need to have emerged to enable DNA transfer across the barrier wall.”
Charles Kurland: We have to remember there’s only a little background of horizontal gene transfer in bacterial populations. The simple reason is that bacteria eat DNA. So sequences are going in all the time. Most of them get chopped up.
Suzan Mazur: Did you say bacteria eat DNA – that’s an interesting way of putting it.
Charles Kurland: Yes, of course. You’re not going to see a lot of DNA left hanging around uneaten. It’s an excellent food. But I agree completely with what Jeff Errington said. We think that the most recent universal common ancestor was the product of a lot of HGT and Carl Woese described the reasons for this.
You see, Ford Doolittle was a postdoc with Carl Woese and he came away with this idea of HGT that Carl in his 1970s papers had described for the progenote -- this very primitive thing. Carl was right on the money there.
We identify MRUCA (most recent universal common ancestor) with the progenote. And I think Jeff Errington is right on the money in that quote you just read about HGT.
If you read Ford Doolittle’s paper of 1999, that paper, which was a very powerful statement of a hypothesis – but in the middle of the paper we are led to forget that it was just a hypothesis. The most important source of data at the time was that gene trees or the evolution of gene sequences was very, very erratic and variable between different genes.
Ford grabbed on that variability as evidence for HGT, and all it is is evidence for variability. The point being that at that time – or at least in Ford’s mind – neutral evolution was a big thing. It was supposed to be based on the assumption of constant rates of mutation. So that variability could not have been -- in Ford’s mind -- due to mutation variability because that was by hypothesis excluded by Kimura and by King and Jukes. So it had to be something else. Ford thought that something else was HGT. It’s all a mirage. We think it’s variable mutation rates.
Suzan Mazur: You also see endosymbiosis as wrong. Would you touch on that a little?
Charles Kurland: Ajith and I are just writing up a paper devoted to that issue. The thing is once you see the phylogeny of the sort that we have, that Ernst Mayr predicted, where the akaryotes are separated in their descent from the eukaryotes, the obvious possibilities for akaryotes to make eukaryotes are just not there. But that’s the essence of the symbiosis hypothesis -- the bacteria gets together with an archaea and it makes a eukaryote, the bacteria becomes the mitochondria and the archaea becomes the nucleus cytoplasmic host. That’s the theory. It’s very, very clearly specified that way.
It’s interesting because it was Lynn’s [Margulis] attempt to explain why DNA was found in the cytoplasm of eukaryotes, and it turned out to be DNA located in mitochondria. It turned out to be DNA located in chloroplasts. Her idea was that: Ah, the mitochondria, the chloroplast, certain kinds of flagella, were actually originally bacteria. Her idea was that if you look at mitochondria, the DNA that was there was the whole genome of that bacteria.
It turns out that it’s nothing like that. And we should have been much more suspicious of the hypothesis as soon as it was discovered that a minor fraction of information for mitochondria is coded in the mitochondria. We should have been very suspicious of that.
Suzan Mazur: She was an amazing presence in science.
Charles Kurland: Yes, she was. When I went for my second postdoc at Berkeley, Lynn was at the Life Sciences laboratory doing her PhD. We got to know one another. We were very friendly. She and her husband at that time, Carl Sagan, who was a tremendous scientist, visited me in Copenhagen when I was a postdoc there. When I went to Berkeley, I kind of hooked up with Lynn. By that time she and Carl Sagan had split up. My wife and I knew her very, very well. She was a force of nature.
Suzan Mazur: I interviewed her at length when she was at Oxford. We had both written for Omni and knew some of the same people going way back. She was, indeed, a force.
But it’s important that science not become dogma. I read some statements you’ve made to that effect, that there’s too much of that going on. I think it’s wonderful that you’re speaking out.
Charles Kurland: It’s definitely a problem. It gets embedded in the education system. There’s no one who takes a biology course that does not believe archaea plus bacteria make eukaryotes. It’s a rule now. It’s beyond criticism now. Well, let’s see if we can change that.
Suzan Mazur: Ajith, you say you were very careful with methods and materials in laying out the ToL. Still things are missing from your analysis, which you and Charles acknowledge in one of your papers. You note linkers that constitute one third to one half of the sequence length of proteins were left out of your computations. Also viruses that move consortially and non-linearly and affect the ToL from bottom to top were left out. So these are two significant missing ingredients. What are your further thoughts, Ajith?
Ajith Harish: Compared to any other ToL, we cover almost 70% of the genome so it is a substantially large amount of data compared to mainstream analysis. Because the technology is not there at the moment, we could not include linkers in our analysis but we plan to do so as the technology improves. Right now we don’t have good analytical methods to find homologous features corresponding to linkers.
Suzan Mazur: The same goes for viruses?
Ajith Harish: Yes, because the annotations are very patchy. Also, because viruses are so small, you cannot really compare them with cellular genomes.
Suzan Mazur: So you’ve gotten as close as you can get for the moment.
Ajith Harish: Yes.
Suzan Mazur: What is some of the resistance you’re encountering to your ToL analysis?
Ajith Harish: First, people were not used to this kind of data, the Superfamily data. The second thing is that we used different kinds of models.
Most models of evolution used to infer the ToL assume that the underlying evolutionary process is stationary and reversible. This implies that the frequencies of characters (traits) do not change during the evolutionary time period and that the changes are unpolarized. As a result they produce unrooted (unpolarized) trees. An unrooted tree has no evolutionary direction: that is from ancestors to descendants.
The standard assumptions of stationary and reversible processes are contradicted by empirical data. That is, the frequencies of characters change during evolution: the process is non-stationary. So when we implement non-stationary models, we cannot only find the root of a tree but can estimate what the ancestral frequencies of different characters are, from the same model. In a way, we can say what the root or starting condition was. We can infer the root using non-stationary models, but not stationary models.
The non-stationary model was not very common, mainly because it’s a bit more sophisticated computationally, mathematically. People didn’t often use these kinds of models before. Now these are available. And in our papers we’ve shown that non-stationary models are better fitting models for the data we have.
Suzan Mazur: Does the discovery of Lokiarchaeaota affect your model?
Charles Kurland: No. The thing that is wonderful about the Loki is that they are big archaea in terms of proteome size and that is attractive for people who want to have an archaea and bacteria becoming a eukaryote. But there’s nothing really substantial by way of phylogeny to support the relationship of that archaea to eukaryotes. In our phylogeny, it’s just one of the archaea.
Suzan Mazur: What are your thoughts about the distinction in cell wall composition in terms of the ToL? Archaea usually have a paracrystalline proteinaceous shell, and bacteria have peptidoglycan walls, either one thick one, or one thin one plus an outer membrane. Eukaryotes have neither.
Charles Kurland: That’s right. They’re all somewhat different.
Suzan Mazur: I was talking to Jeff Errington about this and he said:
“I think it’s curious, really curious that the archaea and the bacteria have a fundamental difference in terms of their cell development structure. It reflects also the very fundamental differences in the way they replicate, transcribe and translate DNA.”
Charles Kurland: Absolutely. There is a split there and in our phylogeny, the split occurs very early. But if you look at the eukaryotes, there are even more early splits. So the plants and the protists and the fungi and the animals – they split out from one another very early in the eukaryotic branch. This business of splitting is something very early. And I agree with Errington that it’s very interesting – what is the basis of this archaeal bacterial split? We can philosophize about it but we have no clear evidence to support any particular way of looking at that. It’s a mystery.
Suzan Mazur: Jeff Errington says the cutting edge is the extent to which bacteria live in the L-form state in a wall-less state. In that state they would proliferate by way of HGT. So how would this affect your ToL?
Charles Kurland: No much at all. Also, I’d like to make a general comment. The cutting edge is in the eye of the beholder.
Suzan Mazur: Jeff was actually referring to the cutting edge of research regarding L-forms.
Ajith Harish: It’s one feature out of so many different features, L-forms. We build a ToL using genomic features, so then it remains to be seen how we can get all these other phenotypic features to correspond to this ToL.
Suzan Mazur: Ajith, did you say the ToL model and methods were part of your PhD – your doctoral dissertation at the University of Illinois, Urbana-Champaign?
Ajith Harish: Partly, yes.
Suzan Mazur: Charles, you say the standard model reflects Aristotelian simple to complex thinking. Your model starts with a complex ancestor and reflects reductive evolution. Would you say more?
Charles Kurland: It gets simpler. This is something that gets very troublesome for people.
Suzan Mazur: In my conversation with Jeff Errington the other day, for instance, he talked about Mycoplasma now being wall-less but it’s related to bacteria that have a wall, so at some point it lost the ability to form a wall. So this is one case of reductive evolution.
Charles Kurland: Yes, of course it is. And there are lots of such examples. The thing that may be behind all of this, the reductive evolution to some degree of simplicity, is that it may be that in the period of time that we’re looking at the environments become simplified and stable. Loss is encouraged by a stable environment.
If you have a toolkit for all kinds of environmental eventualities, and you’re living in an environment that turns out over evolutionary time to be very, very stable – you’re going to start simplifying your toolkit.
Suzan Mazur: That’s right. That’s why as a New Yorker I don’t drive anymore. I’ve lost the ability to drive.
[Laughter]
Charles Kurland: That’s interesting. I don’t drive either. So I think it’s hard to say, but this could be a very direct environmental influence on the evolution of organisms. Environments have become stable and so the organisms can become simpler.
Suzan Mazur: You talked earlier about the problem of science becoming dogma. Can you say a bit more about that?
Charles Kurland: I’m certainly not immune to this because both Ajith and I had to go through some really serious soul searching as the data developed and as our story developed, because both of us entered this game actually quite law abiding citizens of the molecular evolution community and we were essentially trying to solve a technical problem and had no idea that we were getting involved in developing a really different view of evolution.
Suzan Mazur: You’ve gotten some serious support there from the Royal Swedish Science Academy, from the Nobel Committee.
Charles Kurland: Yes. But I think if it were up to most molecular evolutionists, we would be starved to death.
Suzan Mazur: You’ve said that if your thinking about the most recent universal common ancestor being part of an extinct Earthly biosphere is wrong, then the other option is that the origin is extraterrestrial. In light of that thinking, NASA has joined with the Templeton Foundation and the Center of Theological Inquiry to canvas the religious community as to how it would respond to the discovery of extraterrestrial life. Do you have any thoughts about that?
Charles Kurland: Most people would arm to the teeth. As a species we’re very aggressive and very territorial. We are easily frightened. Imagine a monkey colony that has been startled. It would be a very, very scary moment for us to have a spaceship land at Rockefeller Center and – “We come in peace.” We would be shooting at them immediately. And who knows what kind of beings evolved in the Trappist system. They might be nuttier than we are.
Suzan Mazur: NASA and Templeton have thrown almost $3M to this project.
Charles Kurland: I know. I just hope it takes a long time before anyone makes that contact because we have a certain amount of maturation that we have to do before we can handle it. You see what happens with ethnic or racial conflict and migration. Now think what would happen if something really different showed up. . . .
