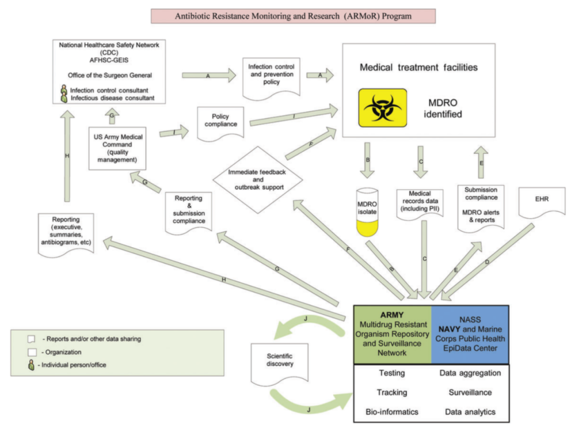
Surveillance and research is key after the first-time discovery of a bacteria carrying the mcr-1 gene - a gene that makes the last-line antibiotic colistin useless against them - in the United States. In May, bacteria carrying the MCR-1 resistance gene were found in human and animal samples for the first time in the United States. Researchers in China had first described findings of the MCR-1 gene in pigs and humans in November 2015. It was since reported in several countries.
Emil Lesho, director and co-founder of the Multidrug-resistant organism Repository and Surveillence Network (MRSN) at Walter Reed Army Institute of Research speaks with us about what needs to be done to prevent a pan-drug resistant superbug. He and his team investigated the case of a 49-year-old woman whose urine contained Escherichia coli harboring the MCR-1 gene.
ResearchGate: How did you find the MCR-1 gene?
Emil Lesho: We found the MCR-1 gene by following the normal operating procedures of our surveillance network. The military has public health mandates that authorize the army and soon all military hospitals to send samples to our central lab.
Normally these are samples of ESKAPE pathogens. The term is an abbreviation of what experts believe are the five most problematic healthcare associated pathogens: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species.
When a lab in a clinical hospital thinks they have found one of those pathogens, they send it to us. They also send us the demographic and clinical information associated with the isolate. We do an extensive array of tests to characterize it. Currently that includes sequencing the genome and antibiotic susceptibility testing and sometimes we also do manual broth dilution. This is a test where we manually dilute an inoculum of the bacteria and confront it with increasing micro-solutions of the antibiotic, to see when and where it responds to it.
In this case, one of our scientists, Patrick Mc Gann was sent a sample by one of the local hospital microbiologists and he happened to be testing these isolates for colistin resistance. That's what alerted him to the possible presence of the MCR-1 gene. He sent us the isolate, we sequenced it and we then confirmed the gene's presence using PCR and by sequencing the whole genome. As part of our normal operating procedure we reported our findings back to the submitting hospital and found that the patient was successfully treated. The good news in this case was that even though the bacteria was resistant to colistin - which is also called the agent of last hope - that didn't mean it was resistant to all antibiotics.
RG: There have been other colistin-resistant bacteria. Why is the MCR-1 gene of such concern?
Lesho: The concern is twofold. Resistance against colistin is usually the result of a mutation caused by existing drug pressure. In the case of MCR-1 gene however, the gene that encodes the resistance sits on a highly mobile genetic element called the plasmid. This plasmid can be easily shared among bacteria of different species. If it were to carry this resistance to colistin to another ESKAPE pathogen then you would potentially end up with an untreatable pathogen that's resistant to all known antibiotics.
Another reason why finding the MCR-1 gene was concerning is that it was found in E.coli. E.coli is the most common cause of urinary tract infection. If the MCR-1 gene became widespread in E.coli, the future scenario for patients with a bladder infection could involve having to go to the hospital for intravenous therapy.
This story does also have two positive elements. For one, as I said, the pathogen was susceptible to a few other antibiotics, including a pill. The other was that it was found in the patient's urine. When bacteria have a high level of resistance and they infect the bloodstream or anatomic sites that are harder for antibiotics to penetrate, like the joints or the spinal fluid, they become very difficult or impossible to treat. In a urinary tract infection however, the antibiotic needs to get to the bladder and urinary tract, where it's present in high concentration by default because the body excretes the medication through the urine.
Lesho: What we know is based on the current literature. It was first found in China in late 2015 and first reported in the medical journal Lancet in February 2016. Since then it has been found in several European countries. The US Department of Agriculture (USDA) also found it in pig intestinal samples. From our preliminary discussions with the USDA, we learned that the plasmid belongs to a different family of plasmids and the bacteria has two different strain types.
As far as the origin of the MCR-1 gene in the sample we investigated is concerned, the Department of Health is currently checking all of the patients' healthcare contacts. The patient didn't have any overseas travel within the last six or seven months and was living a normal life. She isn't an agricultural worker either. We have no clue how she could have been infected with the bacteria carrying this gene. It's not unreasonable to speculate that it will show up more often in the US. Maybe there are isolates that predate the one we found that just haven't been discovered or reported yet.
RG: How are you monitoring the MCR-1 gene?
Lesho: All army hospitals are required to participate with us. Soon this requirement will be extended to the US Navy and Airforce, so that all branches of the service will participate in our network.
We believe in the strength of collaboration and we have partnerships with civilian organizations in nine countries at the moment. We've been contacted by institutions in Kenya, Nigeria and Honduras for help with potential outbreaks of superbugs and wherever possible we try to help with that. We characterize these bacteria for them and do genetic fingerprinting.
We get about 300-600 samples a month and we do extensive testing on all of them which includes sequencing the whole genome. After testing them, we archive the isolates in our repository and the characteristics in our database for future reference. They're then available to researchers who can test assays, vaccines and other countermeasures. Currently we have almost 50,000 of these select ESKAPE pathogens in the repository.
RG: What are the most common types of ESKAPE pathogens in your repository and database?
Lesho: We have thousands of Acinetobacter. Acinetobacter were the pathogens associated with the conflicts in Iraq and Afghanistan. Before the US was involved in those conflicts, the Acinetobacter were fairly susceptible to many antibiotics. It was actually rare to find a highly resistant Acinetobacter. With the severe wounds, polytraumatic injuries and for other reasons - some of them unknown - there was a dramatic increase in highly resistant Acinetobacter.
RG: What research do you think is needed to successfully fight resistance against antibiotics?
Lesho: The challenge these organisms present requires a multi-faceted approach. What we specialize in is surveillance and characterization. You can't overemphasize the importance of surveillance. If you're not surveilling for these pathogens, you don't know what's out there and you don't know if it's getting worse or better. Then, equally important, ongoing surveillance lets you assess what impact or outcome interventions are having. Whole genome sequencing is key in surveillance and fortunately it is becoming faster and more accessible.
The other thing is countermeasure development. People say that as quickly as we can develop a new drug, the bacteria will evolve resistance. We need new drugs, but for many reasons antibiotics development is difficult for big pharma: there are regulatory challenges, there are economic disincentives, and trials also are very expensive and hard to conduct.
In addition to developing new antibiotics, researchers are looking into alternatives. Antibody therapy for instance has been used successfully to treat infections for a while. Other investigators are looking at phage - viruses that attack bacteria. Surveillance, and all of these approaches combined are important to find solutions.
This article originally appeared on ResearchGate News.