Turn on your TV to any crime procedural and there's a good chance you'll see some fancy animation showing some drug molecule, DNA, or protein doing its thing. There will probably be some flashes of light, and the whole thing will be quite dramatic.
Of course, it's also fake. Those images are artists' renderings. Molecules don't really look like colorful balls connected by rigid sticks, and proteins aren't multicolored Pac-Men.
So just where do those images come from?
When, when it comes to proteins and DNA, the answer is mostly X-ray crystallography. In this technique, a crystal of the purified material -- DNA, say, or the protein hemoglobin -- is bombarded by a high-intensity beam of X-rays. These X-rays slice through the crystal, interacting with and bouncing off the regularly spaced atoms in the crystal, producing a complex diffraction pattern that can be used to back-calculate the positions of the atoms in the original crystal.
Think of it like a crime scene investigator working backwards from a spray of bullet holes to figure out where the shooter was standing.
Okay. So, X-ray crystallography tells us what these molecules look like, then, right? Well, not quite. Because X-ray crystallography requires, well, crystals. To make those, researchers take a highly concentrated solution of purified protein and add a chemical that causes the protein molecules to associate with one another. With luck, they will pack themselves in some regular way so that they can form a crystal. The problem is, this step may also force the proteins to assume a shape they don't naturally form in the cell, producing a beautiful, but potentially artificial structure.
And in any event, the resulting structure is the averaged structure of all the molecules in the crystal. According to UCLA researcher Jianwei (John) Miao, that's like averaging the facial features of every person on earth -- you would see that they all have two eyes, two ears, a nose, and a mouth, but all the distinctive variation that makes an individual individual would be blurred away. Similarly, if any one protein in the crystal was just a little bit different from the others, you wouldn't see it -- its signal would be drowned out.
This past week, in the journal Nature, Miao and his colleagues at UCLA and Lawrence Berkeley National Laboratory detailed a method that allows them to view the atoms in nanometer-sized pebbles of gold without averaging. That is, they can view the atoms directly, and on a particle-by-particle basis.
To do that, the researchers used a technique very much like a hospital's CT (or "cat") scan. A CT, or computed tomography scan is like a 3D X-ray. Basically, the machine takes a series of X-ray images of the patient as an X-ray source and detector rotate around the patient's body and then reconstructs those pictures to produce a 3D scan of the patient's insides. In this way, doctors can get a clearer look at what's going on inside the body, to see, say, a tumor that might be hiding beneath another organ.
In this case, Miao and his team used a scanning transmission electron microscope, or STEM, to take 69 pictures -- actually projections produced by an electron beam passing through the material and striking a sensitive detector -- at different angles of a gold particle containing just 23,800 atoms or so. This particle is a "nanoparticle" -- it measures just 10 billionths of a meter in diameter, or about 1/10,000 of the width of a human hair.
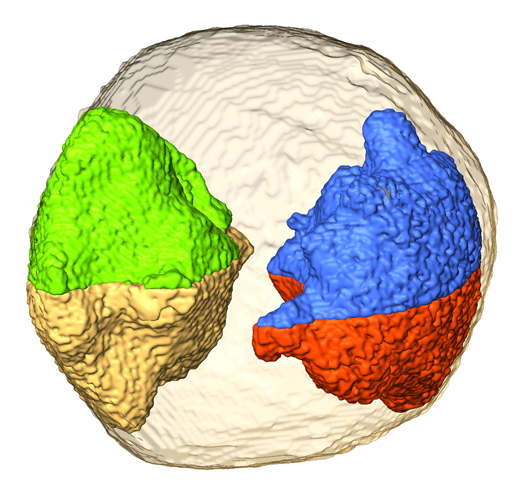
Nanogold in 3D vision. Jianwei Miao and colleagues have developed an electron tomography method to image the 3D structure of a gold nanoparticle at 2.4 Å resolution. Individual atoms are observed in some regions of the particle and several grains are identified at three dimensions. In the figure, four 3D grains (green and gold; blue and red) form two pairs of twin boundaries inside the nanoparticle. This work in principle opens a door for 3D structure characterization of crystalline, polycrystalline and disordered nanomaterials at atomic scale resolution. (Photo credit: Chien-Chun Chen, UCLA)
Armed with their scans, the researchers then crunched the numbers to produce what they believe to be the first atomic scale picture of a single nanoparticle. From the exterior, the particles resemble a slightly squashed icosahedron -- the 20-sided dice of Dungeons & Dragons fame. And that imperfection illustrates the power of Miao's method, he says. Had his team used some sort of averaging experiment (such as X-ray crystallography) that squashing might have been drowned out. But by focusing on just one particle, the team recorded that imperfection in all its golden glory.
The team could even glimpse the "grains" that coalesce to form the intact particle, including their shape and the geometric patterns they form. Not only that, they also got a glimpse of the three-dimensional arrangement of gold atoms on the inside the particles, something that's never been done before, Miao says.
According to Miao, this approach has several potential applications, for instance in the semiconductor industry, where small chemical and structural changes can have profound effects on a material's properties.
One area it won't be useful is in biology -- at least not at the same resolution. The electron beam required to image the particles would destroy fragile biological molecules before they could be imaged. But at lower power, he says, it should be possible to produce lower-resolution, but nonetheless detailed 3D images of the fine structure of cells and viruses.
"We cannot achieve atomic level resolution for biological samples," he says. "But I think we still can do better than any other existing 3D reconstruction method."
The question is, will those new images find their way to your TV screen? Stay tuned.